Krichevsky's research group
In our group we study interactions and physical properties of biological molecules with the help of advanced optical (Single-Molecule Imaging, Fluorescence-Correlation Spectroscopy) and molecular biology techniques.Oleg Krichevsky is a Professor of Physics at Ben Gurion University. He is currently acting as the Physics Department Chairperson; Head of the Steeering Commitee; Head of the Appointments Commitee; and Head of the Office and Lab Space Committee.

Current Projects
Title of Main Project
Sed ut perspiciatis, unde omnis iste natus error sit voluptatem accusantium dolorem que laudantium, totam rem aperiam eaque ipsa, quae ab illo inventore veritatis et quasi architecto beatae vitae dicta sunt, explicabo. Nemo enim ipsam voluptatem, quia voluptas sit, aspernatur aut odit aut fugit, sed quia consequuntur magni dolores eos, qui ratione voluptatem sequi nesciunt, neque porro quisquam est, qui dolorem ipsum, quia dolor sit amet consectetur adipisci[ng] velit, sed quia non numquam [do] eius modi tempora inci[di]dunt, ut labore et dolore magnam aliquam quaerat voluptatem. Ut enim ad minima veniam, quis nostrum[d] exercitationem ullam corporis suscipit laboriosam, nisi ut aliquid ex ea commodi consequatur? [D]Quis autem vel eum i[r]ure reprehenderit, qui in ea voluptate velit esse, quam nihil molestiae consequatur, vel illum, qui dolorem eum fugiat, quo voluptas nulla pariatur? [33] At vero eos et accusamus et iusto odio dignissimos ducimus, qui blanditiis praesentium voluptatum deleniti atque corrupti, quos dolores et quas molestias excepturi sint, obcaecati cupiditate non provident, similique sunt in culpa, qui officia deserunt mollitia animi, id est laborum et dolorum fuga. Et harum quidem reru[d]um facilis est e[r]t expedita distinctio. Nam libero tempore, cum soluta nobis est eligendi optio, cumque nihil impedit, quo minus id, quod maxime placeat facere possimus, omnis voluptas assumenda est, omnis dolor repellend[a]us.
Highlights
- Temporibus autem quibusdam et aut officiis debitis aut rerum necessitatibus saepe eveniet, ut et voluptates repudiandae sint et molestiae non recusandae.
- Tet voluptates repudiandae sint et molestiae non recusandae.
- At vero eos et accusamus et iusto odio dignissimos ducimus, qui blanditiis praesentium voluptatum deleniti atque corrupti
For more information: https://doi.org/123456123456
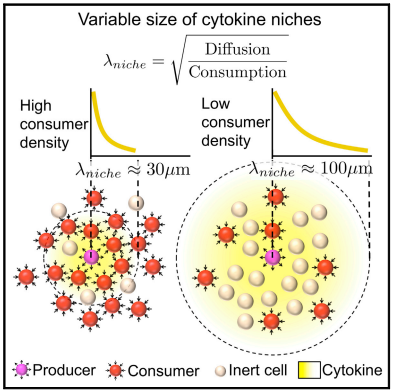
A Tunable Diffusion-Consumption Mechanism of Cytokine Propagation Enables Plasticity in Cell-to-Cell Communication in the Immune System
Immune cells communicate by exchanging cytokines to achieve a context-appropriate response, but the distances over which such communication happens are not known. Here, we used theoretical considerations and experimental models of immune responses in vitro and in vivo to quantify the spatial extent of cytokine communications in dense tissues. We established that competition between cytokine diffusion and consumption generated spatial niches of high cytokine concentrations with sharp boundaries. The size of these self-assembled niches scaled with the density of cytokine-consuming cells, a parameter that gets tuned during immune responses. In vivo, we measured interactions on length scales of 80–120 μm, which resulted in a high degree of cell-to-cell variance in cytokine exposure. Such heterogeneous distributions of cytokines were a source of non-genetic cell-to-cell variability that is often overlooked in single-cell studies. Our findings thus provide a basis for understanding variability in the patterning of immune responses by diffusible factors.
For more information: https://doi.org/10.1016/j.immuni.2017.03.011
Highlights
- Cytokine penetration in tissues is governed by a diffusion-consumption mechanism
- Spherical cytokine niches are generated around cytokine-producing cells
- The characteristic niche size depends on the density of cytokine consumers
- Cytokine niches are a source of variability in otherwise identical cells
Past Research
Internal dynamics of biological polymers: DNA molecules, actin filaments
The problem of polymer dynamics is rather old, going back to the 1930-s. How the stochastic thermal motion (diffusion) reveals itself in the dynamics of polymer segments which are bound by connectivity along the chain, by polymer stiffness, by topological constrains, by hydrodynamic and other interactions? The question does not have simple solutions either in theory, nor in computer simulations, and neither in experiments. We have developed an original experimental approach to measure the dynamics of biological polymers, such as DNA at the level of single monomer with high temporal and spatial resolution.
Furthermore, one does not have to rely on classical thermal fluctuations to drive the dynamics of the system. We introduce now molecular motors “little nanomachines” which actively push polymers around at the nanoscale.
In collaboration with Dr. Anne Bernheim-Groswasser, Chem. Eng.
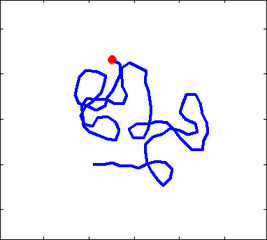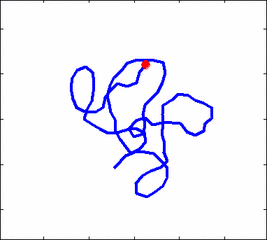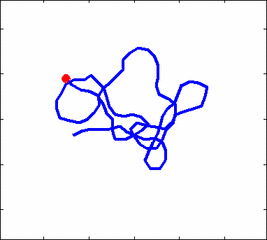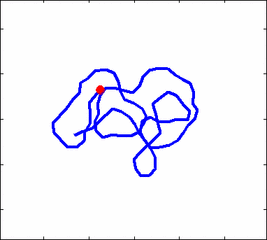
Animation 1: The dynamics of a semi-flexible polymer.
Further reading:
Bacterial nucleoid structure and dynamics and its interaction with the membrane
Bacterial DNA (nucleoid) is highly compacted. Once the nucleoid is gently removed from bacteria, it expands almost 100-fold and occupies the volume many times that of bacteria. What forces keep the nucleoid in its condensed form inside the bacteria? What are the main features of nucleoid structure and dynamics? How nucleoid interacts with the bacterial membrane?
In collaboration with Prof. Itzhak Fishov, Life Sciences.
Further reading:

Figure 1: E. Coli bacteria: the membrane is stained with fluorescent dye.